The Sidra Medicine Core Genomics Lab (SM-CGL) offers access to a wide range of library preparation and sequencing methods. Sidra employs the widely used and industry-leading Illumina Novaseq 6000, Nextseq and Miseq platforms, complemented with Hamilton Robotics automation platforms and cutting-edge analytical instruments for sample quality control. Our wet-lab services are complemented by a standardized sample-level bioinformatics pre-processing pipeline to deliver useable results. Bespoke project-level bioinformatics analysis is available on request through the Genomic Data Science Group.
Whole-Genome Sequencing determines the complete DNA sequence of an organism’s genome and allows the measurement of single nucleotide polymorphisms, copy number variation and detection of small insertions and deletions.
DNA samples submitted to this service are controlled for quantity using a fluorometric DNA quantification assay and for integrity using capillary electrophoresis. Libraries are prepared from 1 µg of DNA using the Illumina TruSeq DNA PCR-free kit with unique dual indices (UDI). After quality control, libraries are sequenced on Illumina Novaseq 6000 instruments to a minimum coverage of 30x human genome (100 Gb, or equivalent) *.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
DNA, eukaryotic species |
|
Concentration: |
> 20ng/µl |
|
|
Amount: |
1 µg in 50 µl |
|
|
Solvent: |
Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/280 > 1.8, average size > 10 kb |
|
|
Labware: |
Thermo Scientific 0.8ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
100 Gb, > 30x human genome* |
|
Quality: |
75% > Q30 |
|
|
File format: |
bam, compressed vcf and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
* multiplexing for smaller genomes or different coverage levels available on request.
** other species’ yield and price depends on genome size, please inquire for full details.
Whole-Genome Sequencing determines the complete DNA sequence of an organism’s genome and allows the measurement of single nucleotide polymorphisms, copy number variation and detection of small insertions and deletions.
DNA samples submitted to this service are controlled for quantity using a fluorometric DNA quantification assay and for integrity using capillary electrophoresis. Libraries are prepared from 300 ng of DNA using the Illumina TruSeq DNA nano kit with unique dual indices (UDI). After quality control, libraries are sequenced on Illumina Novaseq 6000 instruments to a minimum coverage of 30x human genome (100 Gb, or equivalent) *.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
DNA, eukaryotic species |
|
Concentration: |
> 5 ng/µl |
|
|
Amount: |
300 ng in 50 µl |
|
|
Solvent: |
Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/280 > 1.8, average size > 10 kb |
|
|
Labware: |
Thermo Scientific 0.8ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
100 Gb, > 30x human genome* |
|
Quality: |
75% > Q30 |
|
|
File format: |
bam, compressed vcf and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
* multiplexing for smaller genomes or different coverage levels available on request.
** other species’ yield and price depends on genome size, please inquire for full details.
Whole Exome Sequencing uses targeted hybridisation capture to enrich DNA fragments mapping to the expressed regions of the genome, allowing to focus the analysis on variants that directly impact RNA and protein sequences at a much deeper coverage than WGS.
DNA samples submitted to this service are controlled for quantity using a fluorometric DNA quantification assay and for integrity using capillary electrophoresis. Libraries are prepared from 200 ng of DNA using the Agilent SureSelectXT Human All Exon v6 kit covering a 60 Mb target region, with single indices using 12 cycles of PCR for indexing. After quality control, libraries are sequenced on Illumina Novaseq 6000 instruments to the specified coverage. We recommend 100-fold exome coverage for routine samples. At this coverage, we require multiples of 8 samples for multiplexing to offer the most economic cost. For deviating sample numbers, please inquire as the cost might change.
SM-CGL also offer bespoke lower or higher coverages, e.g., 50x for normal samples and 200x for tumour samples. Please inquire for exact pricing.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
DNA, eukaryotic species |
|
Concentration: |
> 20 ng/µl |
|
|
Amount: |
1 µg in 50 µl |
|
|
Solvent: |
Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/280 > 1.8, average size > 10 kb |
|
|
Labware: |
Thermo Scientific 0.8 ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
100x average coverage of Agilent SureSelect XT v6 target region, 12 Gb raw sequence |
|
Quality: |
50% > Q30 and mapping to target region |
|
|
File format: |
bam, compressed vcf and QC report |
|
|
Processing time: |
15 working days when submitting in batches of 8 for up to 8 batches. Does not include wait-to-start time. Inquire for exact details. |
|
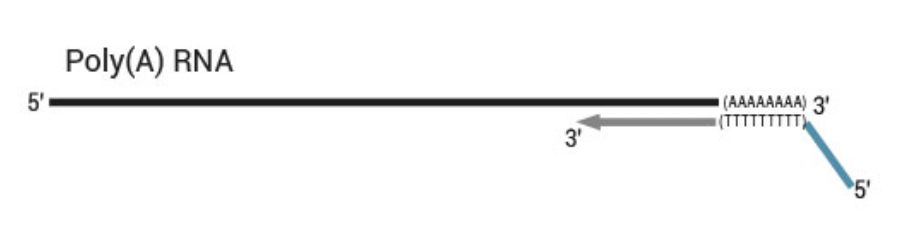
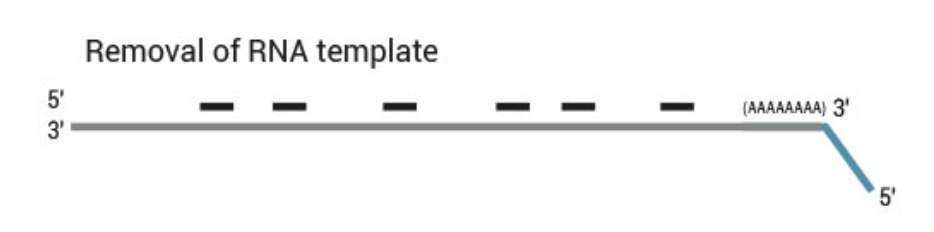
- 3’ differential gene expression sequencing for mRNA
- Only for transcript counting, no splice variants or SNP calling from mRNA
- Requires less sequencing data than traditional mRNA sequencing
- Requires less reads
- Shorter 75 bp SE mode
- Robust towards RNA degradation
- UMIs for PCR duplicate removal
- Globin block for blood samples
- Low input: only 50 ng, less with modifications
Submission criteria:
- 2 Tiers
- Tier 1: > 250 ng in 10 µl (25 ng/µl) [recommended]
- Tier 2: >100 ng in 10 µl (10 ng/µl) > with UMI, more PCR cycles
- RIN > 5
- FFPE possible
- 2 sequencing depth options
- Lexogen 8M: 8M reads, 37 samples per batch
- Lexogen 4M: 4M reads, 75 samples per batch
- All samples run on Nextseq 500 – 75 cycle, single end
RNA sequencing provides insights into the entire transcriptome or messenger RNA (mRNA) profile. Beyond assessing gene expression levels, it can be used to detect transcript isoforms, allele specific gene expression, gene fusions, and expressed single nucleotide variants. Single-stranded RNA is converted into complementary DNAs (cDNA) and double-stranded sequencing libraries, while maintaining the stranded-ness of the measured genes of interest.
mRNA sequencing is a targeted sequencing protocol that enriches for all polyadenylated (poly-A) transcripts and provides gene expression levels, isoform detection and SNV detection. This method is widely used to study gene expression and regulation. By targeting mRNA, only the coding genes are sequenced, which reduces sequencing coverage requirements and maintains a high sensitivity for lowly expressed genes. For blood samples, an optional globin-depletion step is advised.
Total RNA samples submitted to this service are controlled for quantity and quality using the Standard sensitivity RNA assay on the Perkin Elmer Labchip GXII. Libraries are prepared from 500 ng of total RNA using the Illumina TruSeq Stranded mRNA kit. The cDNA obtained after reverse transcription using Superscript IV is ligated with TruSeq RNA Combinatorial Dual Index adapters and amplified for 15 cycles. After quality control, libraries are sequenced on Illumina Novaseq 6000 instruments to a minimum coverage of 19 million reads. Other read depths are available on request.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
RNA |
|
Concentration: |
Minimum 20ng |
|
|
Amount: |
2µg in 50 µl |
|
|
Solvent: |
Rnase free Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/A280 ~2.0, A260/A230 2.0-2.2, RIN > 7 |
|
|
Labware: |
Thermo Scientific 0.8 ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
19 M reads |
|
Quality: |
75% > Q30 |
|
|
File format: |
Bam, read count table and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
Small RNA sequencing (RNA-Seq) is a technique to isolate and sequence small RNA species, such as microRNAs (miRNAs). Small noncoding RNAs act in gene silencing and post-transcriptional regulation of gene expression. Traditional methods for sequencing small RNAs require a large amount of intact total RNA as input, PAGE purification of libraries and these methods used to introduce ligation bias leading to improper representation of small RNA expression. NEXTFLEX® Small RNA-Seq Kit v3 allows completely gel-free library preparation from typical input amounts and it uses randomized adapters reduce ligation bias, resulting in more accurate data.
Total RNA samples submitted to this service are controlled for quantity and quality using the Standard sensitivity RNA assay on the Perkin Elmer Labchip GXII. Libraries are prepared from 250 ng of total RNA using NEXTFLEX® Small RNA-Seq Kit v3. The cDNA obtained after reverse transcription using M-MuLV Reverse Transcriptase is ligated with NEXTFLEX® UDI Barcode adapters and amplified for 16 cycles. After quality control, libraries are sequenced on Illumina Nextseq 550 instrument to a minimum coverage of 4 million reads. Other read depths are available on request.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
Total RNA |
|
Concentration: |
Minimum 250ng |
|
|
Amount: |
2µg in 50 µl |
|
|
Solvent: |
Rnase free Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/A280 ~2.0, A260/A230 2.0-2.2, RIN > 6 |
|
|
Labware: |
Thermo Scientific 0.8 ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
4-8 M reads |
|
Quality: |
75% > Q30 |
|
|
File format: |
Bam, read count table and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
Whole-transcriptome sequencing provides insights into mRNA gene expression as well as the expression of regulatory long non-coding RNAs (lncRNAs), thus providing richer information about regulatory processes in biological systems.
Total RNA samples submitted to this service are controlled for quantity and quality using the standard sensitivity RNA assay on the LabChip GXII. Libraries are prepared from 500 ng of RNA using the Illumina TruSeq Stranded RNA kit. In total RNA sequencing, ribosomal RNA (rRNA) depletion is performed based on the sample type and aims of the project, resulting in depletion of cytoplasmic and mitochondrial ribosomal RNA (human, mouse, rat) and can be combined with a depletion of globin RNA (human, mouse, rat). This depletion is recommended for RNA extracted from whole blood. cDNA obtained after reverse transcription using Superscript IV is ligated with TruSeq RNA Combinatorial Dual Index adapters and amplified for 15 cycles. After quality control, libraries are sequenced on Illumina Novaseq 6000 instruments to a minimum coverage of 50 million reads. Higher read depths are available on request.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
RNA |
|
Concentration: |
> 50 ng/µl |
|
|
Amount: |
2µg in 50 µl |
|
|
Solvent: |
RNAse-free Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
A260/A280 should be ~2.0, A260/A230 should be 2.0-2.2, RIN > 7 |
|
|
Labware: |
Thermo Scientific 0.8 ml 96-well storage plate (AB-0765) |
|
|
OUTPUT |
Yield: |
50 M reads |
|
Quality: |
75% > Q30 |
|
|
File format: |
Bam, read count table and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
Single cell sequencing technologies provide a high-resolution view of complex biological systems and processes, revealing molecular mechanisms of development and disease at the level of individual cells. Direct analysis of tissues or cellular populations at the single cell level allows to characterize intracellular heterogeneity, cell types and states, and dynamic cellular transitions.
Libraries are constructed with Next GEM Single Cell Gene Expression v3.1 kit from studied cell cultures. Libraries are sequenced using on a NovaSeq 6000 with a depth of > 50,000 raw reads per cell, with raw sequencing data analyzed and visualized with Cell Ranger software.
We strongly encourage users to refer to 10X Genomics recommendations and publications and discuss their project and sample types before using this service.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
Cell culture |
|
Concentration: |
700-1,200 cells/uL and the viability should be >90% |
|
|
Amount: |
50-100 µl |
|
|
Solvent: |
1X PBS with 0.04% BSA (Cell suspension buffers should be free of EDTA and Mg++ as well as free of DNAse) |
|
|
Quality: |
Viability of cells should be 80 - 90% and cells must be counted accurate instrument before submission |
|
|
Labware: |
Eppendorf Tubes |
|
|
OUTPUT |
Yield: |
250 M per sample |
|
Quality: |
75% > Q30 |
|
|
File format: |
Bam, read count table and QC report |
|
|
Processing time: |
10 working days when submitting in batches of 8 for up to 20 batches. Does not include wait-to-start time. Inquire for exact details. |
|
Sequencing of user-prepared libraries and library pools on Illumina Novaseq 6000 and Nextseq 500 instruments is a way to enable cost-efficient data generation for library types that are not offered routinely. SM-CGL offer the popular 150 bp paired-end mode to all users. Other read modes (75 bp PE, single-end modes) are available on request and subject to demand, please inquire.
Users are required to take responsibility for generating suitable, high-quality library types and, if applicable, create pools with a balanced index representation to avoid data quality issues. SM-CGL will quality control the submitted libraries and/or library pools using the 2100 Agilent Bioanalyzer and qPCR to assess the library/pool concentration and size distribution and can only advise based on these parameters, as well as the selected indices. We strongly encourage users to discuss their project and library types before using this service.
|
SPECIFICATIONS |
||
|
INPUT |
Input material: |
Illumina-compatible library pool |
|
Concentration: |
|
|
|
Amount: |
3 nM in 20 ul |
|
|
Solvent: |
Water, Tris-EDTA, Qiagen EB |
|
|
Quality: |
350 < x < 900 bp |
|
|
Labware: |
Eppendorf LoBind 1.5 ml tube, fully skirted PCR plate |
|
|
OUTPUT |
Yield: |
Depending on instrument choice |
|
Quality: |
75% > Q30 |
|
|
File format: |
Compressed Fastq |
|
|
Processing time: |
15 working days when submitting in batches of 8 for up to 8 batches. Does not include wait-to-start time. Inquire for exact details. |
|